The blogdown package is now on CRAN. Read on for highlights from the version 1.0 release, including smoother workflows, new checking functions to guide you into the pit of success, the ability to pin Hugo versions, better organization of content files via page bundles, and the new Markdown mode for R Markdown posts. Blogdown: The 'blogdown' package builddir: Build all Rmd files under a directory buildsite: Build a website bundlesite: Convert post files to leaf bundles checksite: Provide diagnostics for a website project cleanduplicates: Clean duplicated output files confignetlify: Create the configuration (file) for Netlify configRprofile: Create or modify the '.Rprofile' file for a website project.
R Markdown (Allaire et al. 2021) is a plain-text document format consisting of two components: R (or other computing languages) and Markdown. Markdown makes it easy for authors to write a document due to its simple syntax. Program code (such as R code) can be embedded in a source Markdown document to generate an output document directly: when compiling the source document, the program code will be executed and its output will be intermingled with the Markdown text.
R Markdown files usually use the filename extension .Rmd. Below is a minimal example:
Such a document can be compiled using the function rmarkdown::render(), or equivalently, by clicking the Knit button in RStudio. Under the hood, an R Markdown document is first compiled to Markdown through knitr(Xie 2021), which executes all program code in the document. Then the Markdown output document is compiled to the final output document through Pandoc, such as an HTML page, a PDF document, a Word document, and so on. It is important to know this two-step process, otherwise you may not know which package documentation to look up when you have questions. Basically, for anything related to the (R) code chunks, consult the knitr documentation (https://yihui.org/knitr/); for anything related to Markdown, consult the Pandoc documentation (https://pandoc.org).
An R Markdown document typically consists of YAML metadata (optional) and the document body. YAML metadata are written between a pair of --- to set some attributes of the document, such as the title, author, and date, etc. In the document body, you can mix code chunks and narratives. A code block starts with a chunk header ```{r} and ends with ```. There are many possible chunk options that you can set in the chunk header to control the output, e.g., you can set the figure height to 4 inches using ```{r fig.height=4}. For all possible chunk options, see https://yihui.org/knitr/options/.
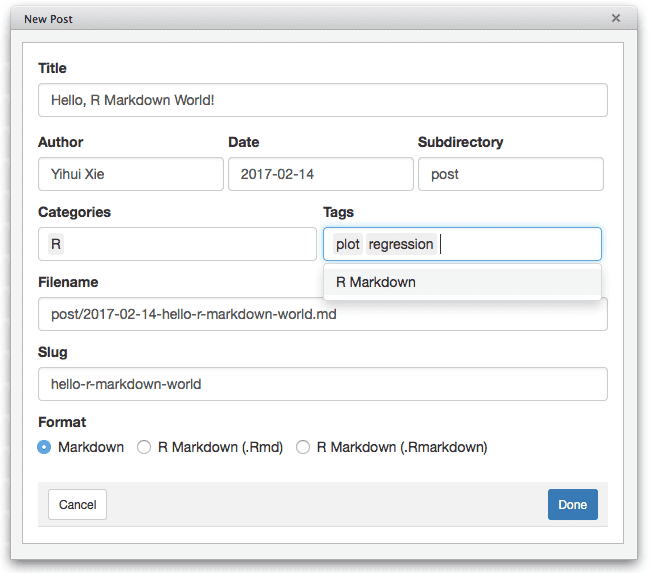
Pandoc supports a large variety of output document formats. For blogdown, the output format is set to HTML (blogdown::html_page), since a website typically consists of HTML pages. If you want other formats, please see Section 2.7. To create an R Markdown post for blogdown, it is recommended that you use the RStudio “New Post” (Figure 1.2) or the function blogdown::new_post(), instead of the RStudio menu File -> New File -> R Markdown.
You are strongly recommended to go through the documentation of knitr chunk options and Pandoc’s manual at least once to have an idea of all possibilities. The basics of Markdown are simple enough, but there are many less well-known features in Pandoc’s Markdown, too. As we mentioned in Section 1.5, blogdown’s output format is based on bookdown(Xie 2020a), which contains several other Markdown extensions, such as numbered equations and theorem environments, and you need to read Chapter 2 of the bookdown book (Xie 2016) to learn more about these features.
You can find an R Markdown cheat sheet and a reference guide at https://www.rstudio.com/resources/cheatsheets/, which can be handy after you are more familiar with R Markdown.
With R Markdown, you only need to maintain the source documents; all output pages can be automatically generated from source documents. This makes it much easier to maintain a website, especially when the website is related to data analysis or statistical computing and graphics. When the source code is updated (e.g., the model or data is changed), your web pages can be updated accordingly and automatically. There is no need to run the code separately and cut-and-paste again. Besides the convenience, you gain reproducibility at the same time.
 5 min read2017/05/10
5 min read2017/05/10Blogdown is an R package that allows people to write web sites, especially weblogs, including R code and its output.
By default, blogdown uses a programme called Hugo, which converts Markdown files into static web sites. This is a bit like Jekyll, the software behind GitHub Pages.
What blogdown adds is the ability to write R Markdown posts. These are converted directly into HTML via the rmarkdown R package, which itself uses pandoc.
The writing process works like this1:
As you can see, you have the choice of writing your posts in R Markdown, or, assuming you aren’t including any executed R code, in Markdown directly. However, Hugo’s own flavour of Markdown, “Blackfriday”, is different to R Markdown syntax, which is based on pandoc. This can get very confusing quite quickly, so just write everything in R Markdown. If you don’t need to run R chunks, simply don’t include any in your .Rmd file.
Installation
Blogdown and Hugo are easy to install on Windows, unlike Jekyll, which is written in Ruby and generally a pain in the arse. To install blogdown, run
whence you can install Hugo directly from within R:
Create a new site
In RStudio, go File > New Project… > New Directory > Empty Project. Then run
and everything you need to get started will magically appear!
Write a post
Everything works exactly like a regular R Markdown document, except a few options in the YAML header. Some sample posts are generated when you first create the site, so you can try editing these.
To create a new post, run
or simply create an .Rmd document from scratch in the content/post/ folder. So that everything appears in the right order in a blog feed, file names should be prefixed with the publication date, in the format YYYY-MM-DD-. You will see in this folder that some posts are in plain Markdown as .md files and processed by Hugo. Files written in the .Rmd format (probably everything you are likely to write) are converted into html by blogdown and stored here, then served by Hugo.
Publish to GitHub Pages
Some subtleties here, as the blogdown documentation is incomplete. Rather than GitHub Pages, blogdown’s authors recommend alternative services such as Netlify or Updog, but I have not tried those yet.
Blogdown R Examples
To begin, create a GitHub account if you do not already have one. GitHub Pages sites appear on the web at the address username.github.io/repository_name. You will also need to install git on your computer: download it from git-scm.com, install it and make sure it is on your PATH.
Now, in theory we can only track changes in the generated site, which would involve initialising a Git repository in your blogdown site directory’s public/ folder. This seems silly to me: we never edit the HTML files directly so surely we want to keep track of the source code as well.
In this case, create a new repository on GitHub and call it whatever you like. Go into the repository settings and scroll down to where it says “GitHub Pages”. Click on the dropdown box, underneath where it says “Source”. Select master branch /docs folder. This means it will serve up a web site from your content, but only that stored in the docs/ subfolder. Everything else will be tracked for version control, however.
Of course, everything is actually being served into the public/ folder, so we need to fix that. Open config.toml and add/modify the top of the file to include this line:
Now when you regenerate the site using
Blogdown R

or
it should put everything in docs/ instead of public/. You can delete the public/ folder now as it is no longer needed.
In the main directory, create an empty file called .nojekyll. You can do this in R using file.create('.nojekyll'). This stops GitHub from trying to process your site using Jekyll; when you upload it, it will already have been generated by Hugo.
Now, back in RStudio, go to Tools > Project Options… > Git/SVN. At the “Version control system” dropdown box, select “Git”. A dialogue box will ask if you would like to initialise a new Git repository. Say “yes”.
You should get a tab appearing in the top-right of your RStudio session that says “Git”. Open this pane and tick all the boxes next to the files (or docs/ and .nojekyll at the very least). Click “Commit” and enter a suitably descriptive message (“Initial commit” is popular for your first one), click “Commit” and close the pop-up dialogues once everything has completed.
We want to connect our local git repository with the one on GitHub. To do that, in your Git tab, click More > Shell…. A command prompt (terminal) will open. Don’t panic; just enter the following lines (which will be suggested to you on your GitHub repository page):
replacing USERNAME with your own GitHub username and REPONAME with the name of your repository. When prompted, enter your GitHub username and/or password (you can avoid being prompted for your username by connecting via SSH).
Close the terminal—you shouldn’t been needing it again.
Wait a few minutes and then visit http://USERNAME.github.io/REPONAME. What do you see?
In future, you can commit and then push changes to the Web using the “Push” button in RStudio.
Blogdown Book
Incidentally, this flow diagram was created using DiagrammerR.↩
